Sharing Phylogenetic Data — Public Comment Invited
Data sharing is important–it helps scientists to reproduce others’ results, add data to previous analyses, and otherwise maximize the impact of an individual publication. This isn’t really news, of course. But, now that we are in a world where data sharing is increasingly the norm, how do we make sharing as easy as possible? It’s not enough to slap some tables up on a personal website or even on an official data repository; the data have to be easily readable by humans and machines, easily reusable, and easily accessible for the long term.
One major goal of biology is to understand the arrangement of the branches on the Tree of Life (phylogenetics). Beyond the general drive to understand what is most closely related to what, a solid tree can help answer all sorts of interesting questions about the patterns and process of evolution. How did gigantism evolve in some dinosaurs? How did bird brains evolve? How did bone-cracking dogs get their beefy skulls? Of course, published trees are “just” hypotheses. The arrangement changes slightly (or occasionally even radically) with the discovery of new species, sequencing of new genomes, development of new tree reconstruction methods, and reinterpretation of old anatomy. Thus, data sharing is crucial for phylogenetics–scientists need to be able to add data, reanalyze old data sets, and reevaluate previous work in order to have the best possible tree.
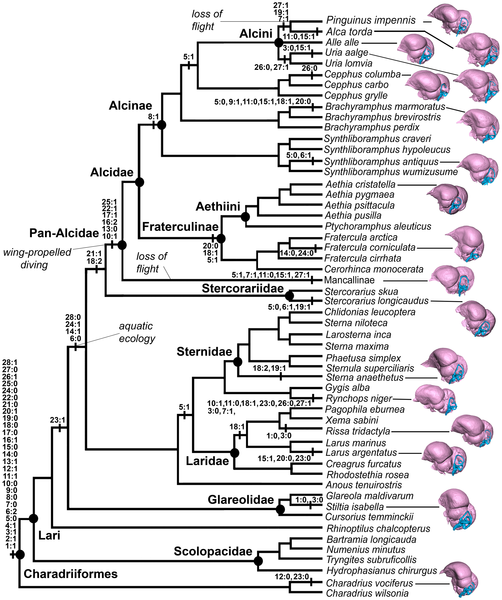
Data tables form the core of phylogenetic analyses, whether the data are in the ACGT of a DNA sequence or the 1’s and 0’s that mark presence or absence for the anatomy of a fossil organism. Alongside the data table are lists of characters, lists of specimens, and all of the other important supporting information. This makes for a complex situation, and there is considerable variability in how different researchers, and even different publishers, present these critical data. Standardization is a must!
Karen Cranston, Luke Harmon and Maureen O’Leary, lead researchers on the NSF-funded “Assembling, Visualizing, and Analyzing the Tree of Life” (AVAToL) project, are drafting a document that lays out best practices for sharing phylogenetic data. This includes trees, aligned gene sequences, data matrices, character lists, and all of the other technical details that underpin an analysis. Maureen relayed to me that she is particularly interested in comments on how to deal with time and other metadata. Are you someone who works with or is interested in phylogenetic data? They’re looking for your input!
The draft “best practices” guidelines are available as a Google Document, freely editable by anyone. In particular, input is requested on guidelines for data storage and formatting. The coordinators for this effort request your comments soon—January 22 or 23 at the latest, so act now if you are interested! There are already some great comments on the document.
I applaud the steps that Cranston, Harmon, and O’Leary are taking to make this a true community effort. Too often these kinds of “best practices” documents are crafted in the proverbial “smoke-filled back room,” catering to a handful of insiders. The more open the process, the better the guidelines and the more likely people are to use them! May this serve as a model for future efforts.
Want to add your input? Check out the draft guidelines for more information!
